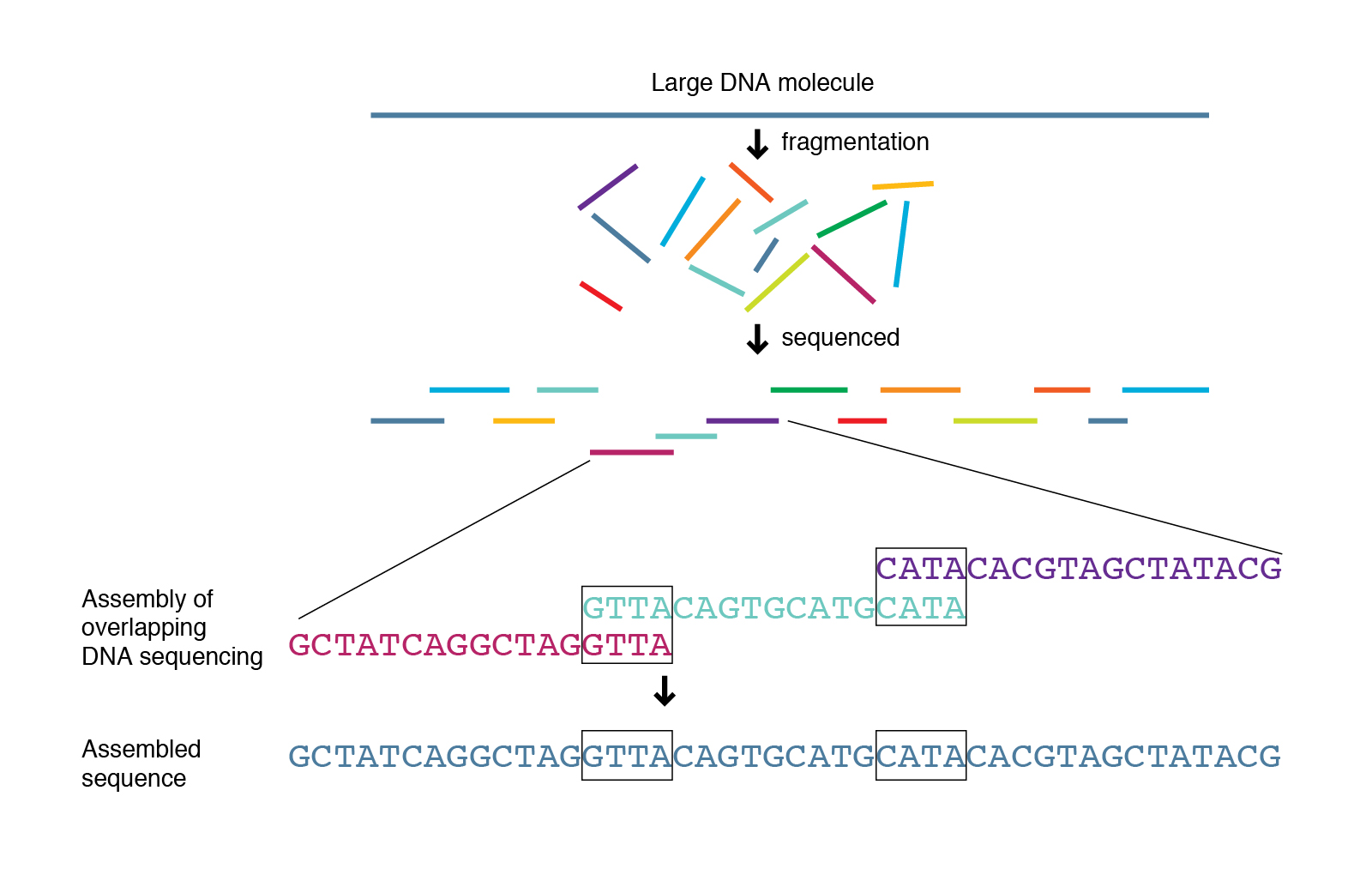
paired end sequencing wikipedia
Learn what Next-Generation Sequencing NGS technology is and what it means. Illumina your reads are supposed to align FR and if they instead align RF FF or RR thats a problem and often indicates the reads aligned incorrectly though it could also mean they aligned correctly and that a real inversion or translocation exists in the samples genome see notes from Devin Abshers.

Continents And Oceans Ks1 Lesson Plan Activities Teaching Resources Continents And Oceans World Map Continents Continents
Learn about emulsion PCR ePCR one of the PCR techniques used in next-generation sequencing.

. The second notation is. BioWare Main_Page Trimmomatic performs a variety of useful trimming tasks for illumina paired-end and single ended dataThe selection of trimming steps and their associated parameters are supplied on the command line. Illumina sequencing and array technologies fuel advancements in life science research translational and consumer genomics and molecular diagnostics.
In DNA sequencing lingo the words paired-end PE and mate-pair MP are frequently used interchangeably. Typical experimental design advice for expression analyses using RNA-seq generally assumes that single-end reads provide robust gene-level expression estimates in a cost-effective manner and that the additional benefits obtained from paired-end sequencing are not worth the additional cost. These fragments are.
2 For paired-end RNA-Seq use the following kits with an alternate fragmentation protocol followed by standard Illumina paired-end cluster generation and sequencing. This can be very helpful e. Paired-end libraries are created like regular fragment libraries but they have adaptor tags on both ends of the DNA insert that enable sequencing from two directions.
Cutadapt FastX_toolkit PrinSeq Seq_crumbs. For your De novo genome assembly Fig. For those not familiar with paired-end reads check out this post.
A flexible read trimming tool for Illumina NGS data. Since the beginning of 2013 this preparation has been based on Nextera. Combining data from mate pair sequencing with those from short-insert paired-end reads provides increased information for maximising sequencing coverage across a genome 1.
The figure shows the workflow for mate-pair library preparation for Illumina sequencing. 500 bps gives you two sequences. Thus far we have learned how to sequence DNA using.
However in many cases eg with Illumina NextSeq and NovaSeq. Paired-end RNA sequencing RNA-Seq enables discovery applications such as detecting gene fusions in cancer and characterizing novel splice isoforms. There is a unique adapter sequence on both ends of the paired-end read labeled Read 1 Adapter and Read 2 Adapter.
1 shows a schematic view of an Illumina paired-end read. But if youre just doing conventional paired-end sequencing ie. Now lets get started.
QC Metric Guidelines mRNA total RNA RNA Types Coding Coding non-coding RIN 8 low RIN 3 bias 8 Single-end vs Paired-end Paired-end Paired-end Recommended Sequencing Depth 10-20M PE reads 25-60M PE reads FastQC Q30 70 Q30 70 Percent Aligned to Reference 70 65 Million Reads Aligned Reference 7M PE reads or 14M reads 165M PE reads. First of all I checked if Sequence Id contains paired end notation. Learn about how Next-Generation Sequencing techniques are used today to rapidly sequence billions of DNA base pairs for low costs.
One big amplicon of eg. Otherwise they can be single end. Paired-end libraries allow users to sequence the DNA fragment from both ends instead of typical sequencing which occurs only in a single direction.
As described in this wikipedia page for Illumina reads there are two possible notation for singlepaired end reads. When you are sequencing paired-end then you are sequencing one time forward barcode adapter the sequence you want to have and one time reverse barcode adapter the sequence you want to have. 1 DNASequencingOverviewRecap 2 Templatepreparation 3 Sequencing-by-synthesis 4 Singleandpaired-endreads 5 References F.
Figure 4Paired-EndSequencingandAlignmentPaired-endsequencingenablesbothendsoftheDNAfragmenttobesequencedBecausethedistancebetweeneach pairedreadisknown. Mate pair sequencing is used for various applications applications including. The forward sequence also called forward read and the reverse read.
For more detailed analyses to determine for example allele-specific expression or expression of low-abundant transcripts 60 million to 100 million reads may be required. For the first test I took some sequence from the human genome hg19 and created two 100 bp reads from this region. Learn about the difference between Paired-End and Single-Run sequencing and why the former creates more precise alignments than the latter especiall.
Dündar ABC WCM Illuminas sequencing by synthesis January 21 2020 2 38. Read 1 often called the forward read extends from the Read 1 Adapter in the 5 3 direction towards Read 2 along the forward DNA strand. The preparation of mate pair libraries is designed to allow classical paired-end sequencing of both ends of a fragment with an original size of several kilobases.
While the underlying principles between PE and MP reads have strong similarities there are inherent differences that are crucial to understand. It has very nice and simple illustrations along with explanations on the terminology used in paired-end sequencing. For sequencing projects that require higher accuracy such as studies of alternate splicing 40 million to 60 million paired-end reads will provide better results.
If the last number is 2 in some reads then the reads are paired end. Paired-end tags PET sometimes Paired-End diTags or simply ditags are the short sequences at the 5 and 3 ends of a DNA fragment which are unique enough that they theoretically exist together only once in a genome therefore making the sequence of the DNA in between them available upon search if full-genome sequence data is available or upon further. Paired end sequencing refers to the fact that the fragment s sequenced were sequenced from both ends and not just the one as was true for first generation sequencing.
For the above protocol we have developed a novel algorithm ePEST ChIP-exo paired-end sequencing processing toolkit which leverages on the statistical powerful of r-scan for detecting binding peaks with son-3-end reads and Chernoff inequality for identifying precise borders with exo-5-end reads respectively.

Pin On Teaching Asian American P I History
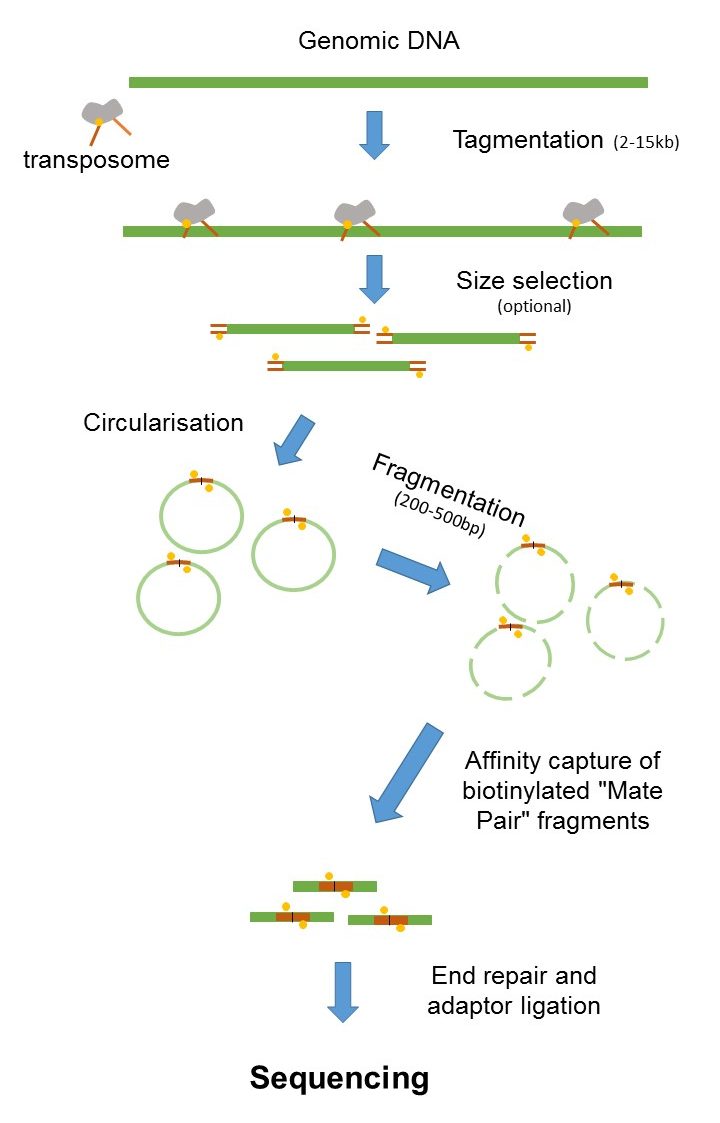
Mate Pair Sequencing France Genomique

Paired End Shotgun Sequencing Okinawa Institute Of Science And Technology Graduate University Oist

What Is Mate Pair Sequencing For

Illumina Paired End Sequencing Youtube

What Is The Difference Between Single End And Paired End In Sequencing
Difference Between Whole Genome Sequencing And Next Generation Sequencing Difference Between

What Are Paired End Reads The Sequencing Center

Bacterial Chromosome Replication Chromosome Biology Microbiology

What Are Paired End Reads The Sequencing Center
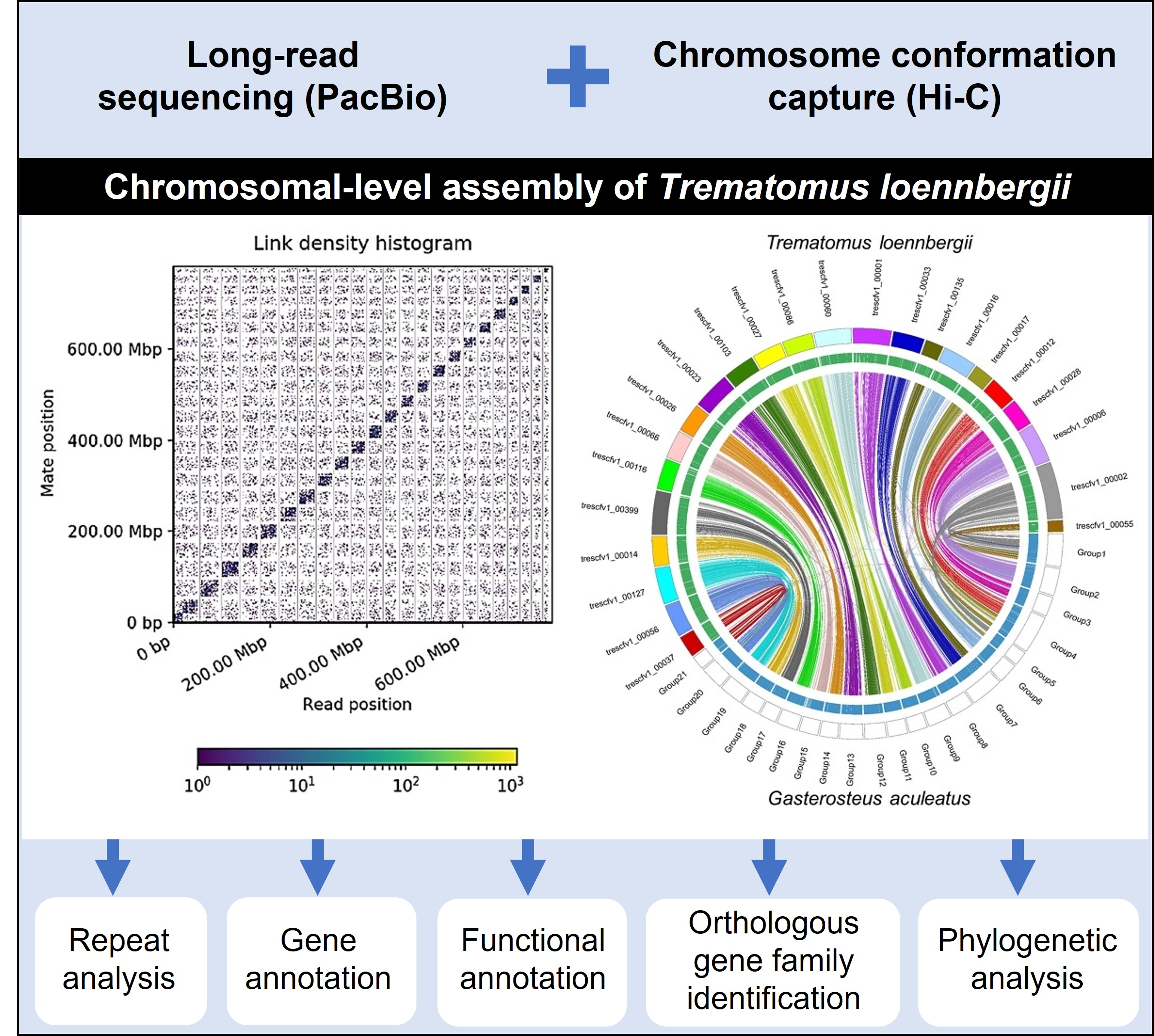
Diversity Free Full Text Chromosomal Level Assembly Of Antarctic Scaly Rockcod Trematomus Loennbergii Genome Using Long Read Sequencing And Chromosome Conformation Capture Hi C Technologies Html
Plos Computational Biology Informatics For Rna Sequencing A Web Resource For Analysis On The Cloud

What Is Mate Pair Sequencing For

Bacterial Chromosome Replication Chromosome Biology Microbiology